Welch Two Sample t-test
data: control and treatment
t = -0.58009, df = 194.18, p-value = 0.5625
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-0.3519980 0.1919951
sample estimates:
mean of x mean of y
0.03251482 0.11251629 Interlude: Correlated measures
Power analysis through simulation in R
Niklas Johannes
Takeaways
- Understand difference between independent and correlated measures
- Know how to simulate correlated measures
- Get to know the
whilelogic
So far
So far we’ve been drawing samples from two independent groups.
What about two measures from the same person
- Think of a typical pre/posttest design
- Someone who has a tendency to score low will therefore score low on both pre- and post-test measure
- The measures are correlated
- We need to take into account that measures come from the same unit when simulating
So how do we get correlated measures?
- We need to increase the dimensions
- So far, we’ve worked with one dimension: our dependent variable only
- But if a person has multiple measures, that means we don’t just have one normal distribution
- We have two correlated normal distributions: a multivariate normal distribution
How does that look like?
For univariate, we pick from a single value (left). For bivariate, we pick two values, or a point on the the plane (right).
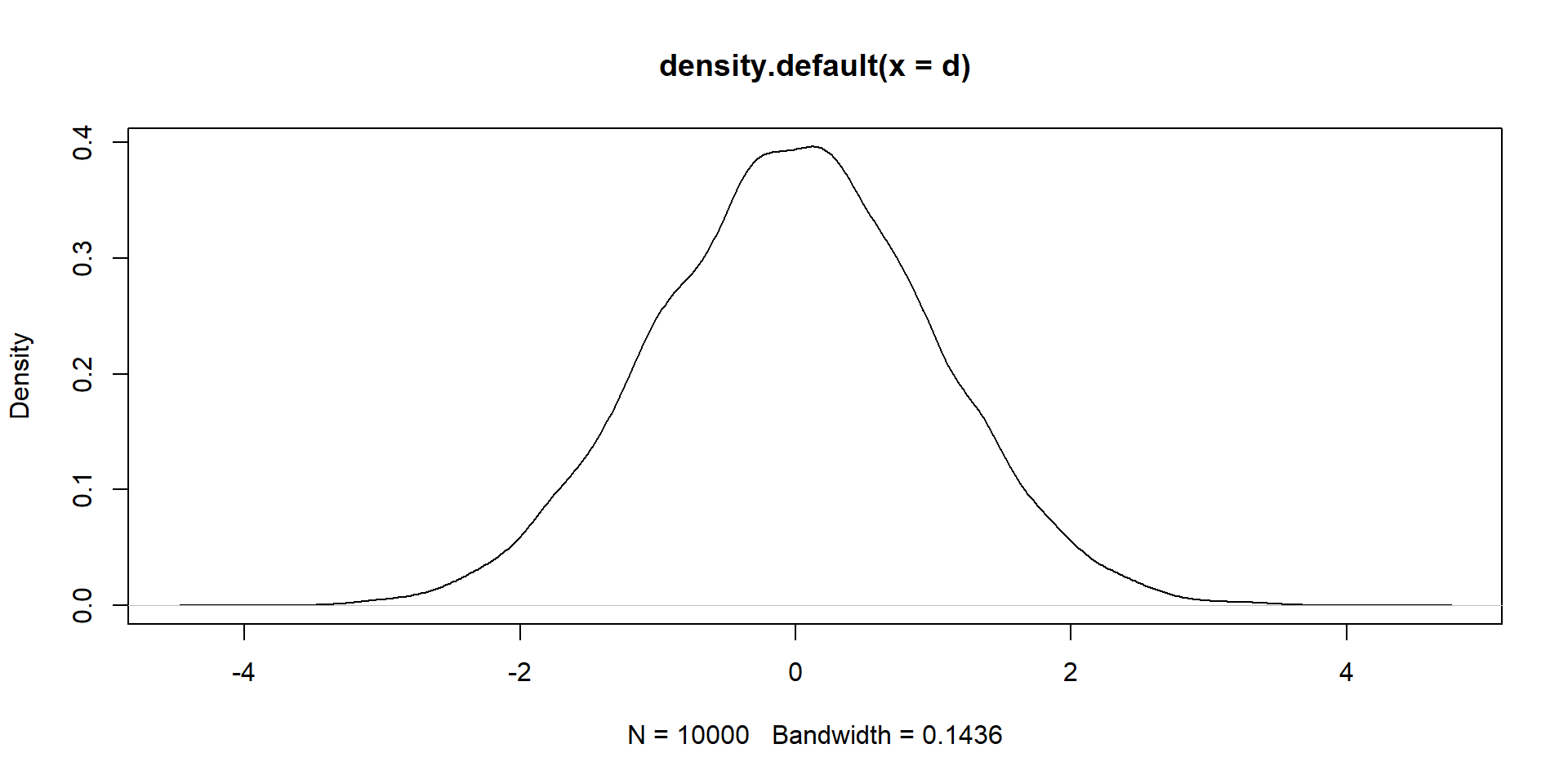
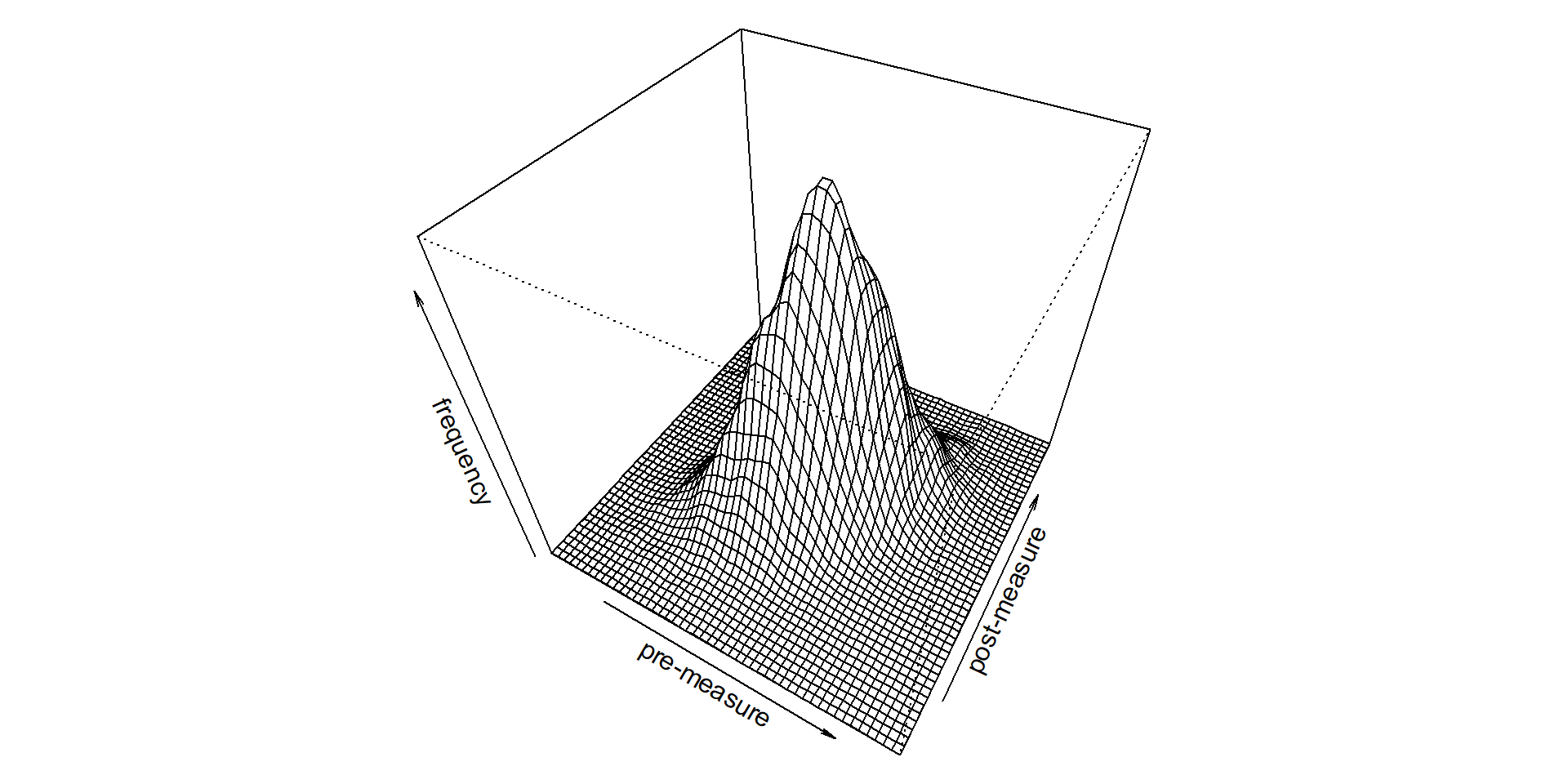
For the left, we need a mean and SD. What do we need for the right?
What goes into a multivariate distribution
Everything’s double:
- 2 means
- 2 SDs
- Correlation between variables
- An SD for the entire “mountain”
SD = Variance-covariance matrix
The SD for the “mountain” is just the SDs and correlations between the two variables in one place so that we can draw our data from them.
\[ \begin{bmatrix} var & cov \\ cov & var \\ \end{bmatrix} \]
This isn’t new
All of you have done correlation tables: they’re just standardized versions of the variance-covariance matrix.
\[ \begin{bmatrix} SD & r \\ r & SD \\ \end{bmatrix} = \begin{bmatrix} 1 & 0.5 \\ 0.5 & 1 \\ \end{bmatrix} \]
So how do we make this?
\[ \begin{bmatrix} var & cov \\ cov & var \\ \end{bmatrix} = \begin{bmatrix} ? & ? \\ ? & ? \\ \end{bmatrix} \]
Our values
Say we have an experiment where people give us a baseline measure, then the treatment happens, and we get a post-treatment measures. The measures are normally distributed with means of 10 and 10.5 and SDs of 1.5 and 2. The pre- and post-measure are correlated with \(r\) = 0.4.
Getting variance and covariances
SD is just the square root of the variance. So we go \(Var = sd^2\) and we got our variance.
Covariance is just the correlation times the SDs. So we go \(covariance = r(pre, post) \times sd_{pre} \times sd_{post}\)
Now let’s combine all that into a matrix
Ready to simulate now
We use the mvrnorm function for, well, multivariate normal distributions, from the MASS package. Let’s get a massive sample of 10,000 people.
Let’s check that
Let’s first check the sample to see whether we can recover our numbers.
Run our test
The while operator
Logic
At this point, we’ve worked with for loops and went from a minimum to a maximum. If that maximum is large, that can take quite some time. You can also consider the while function to stop when you’ve reached the point you want to be at.
Easy example
How would that work for our purposes?
draws <- 1e3
n <- 60
effect_size <- 0.5
d <- data.frame(
sample_size = NULL,
power = NULL
)
power <- 0
while (power<0.95) {
pvalues <- NULL
for (i in 1:draws) {
control <- rnorm(n)
treatment <- rnorm(n, effect_size)
t <- t.test(control, treatment, alternative = "less")
pvalues[i] <- t$p.value
}
power <- sum(pvalues<0.05) / length(pvalues)
d <- rbind(
d,
data.frame(
sample_size = n,
power = power
)
)
n <- n + 1
}What did we just do?
sample_size power
1 60 0.845
2 61 0.856
3 62 0.872
4 63 0.854
5 64 0.885
6 65 0.874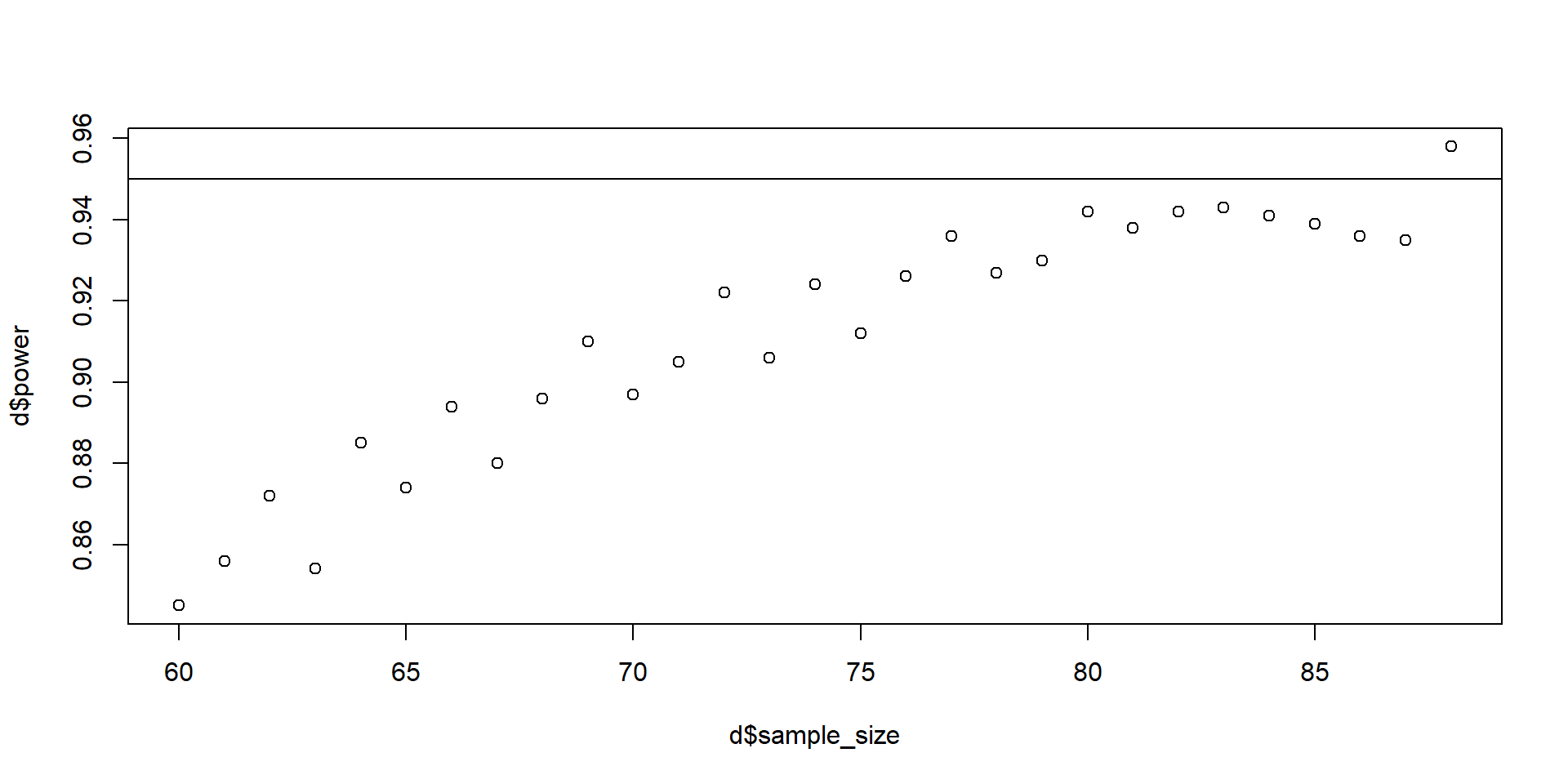
Takeaways
- Understand difference between independent and correlated measures
- Know how to simulate correlated measures
- Get to know the
whilelogic
