The linear model and comparing multiple groups
Power analysis through simulation in R
Niklas Johannes
Takeaways
- Understand the logic behind the data generating process
- See how the linear model is our data generating process
- Apply this to a setting with multiple categories in a predictor
Lots of foreshadowing
- Data generating process
- Simulations are that process
What’s that data generating process?
- The process, in the real world, that you believe created the data
- We don’t know the process, but we can observe the outcomes
- When we try to explain the outcome, we make assumptions about the data generating process
- We collect data from samples and want to know whether our assumptions fit (and generalize)
What’s the process?
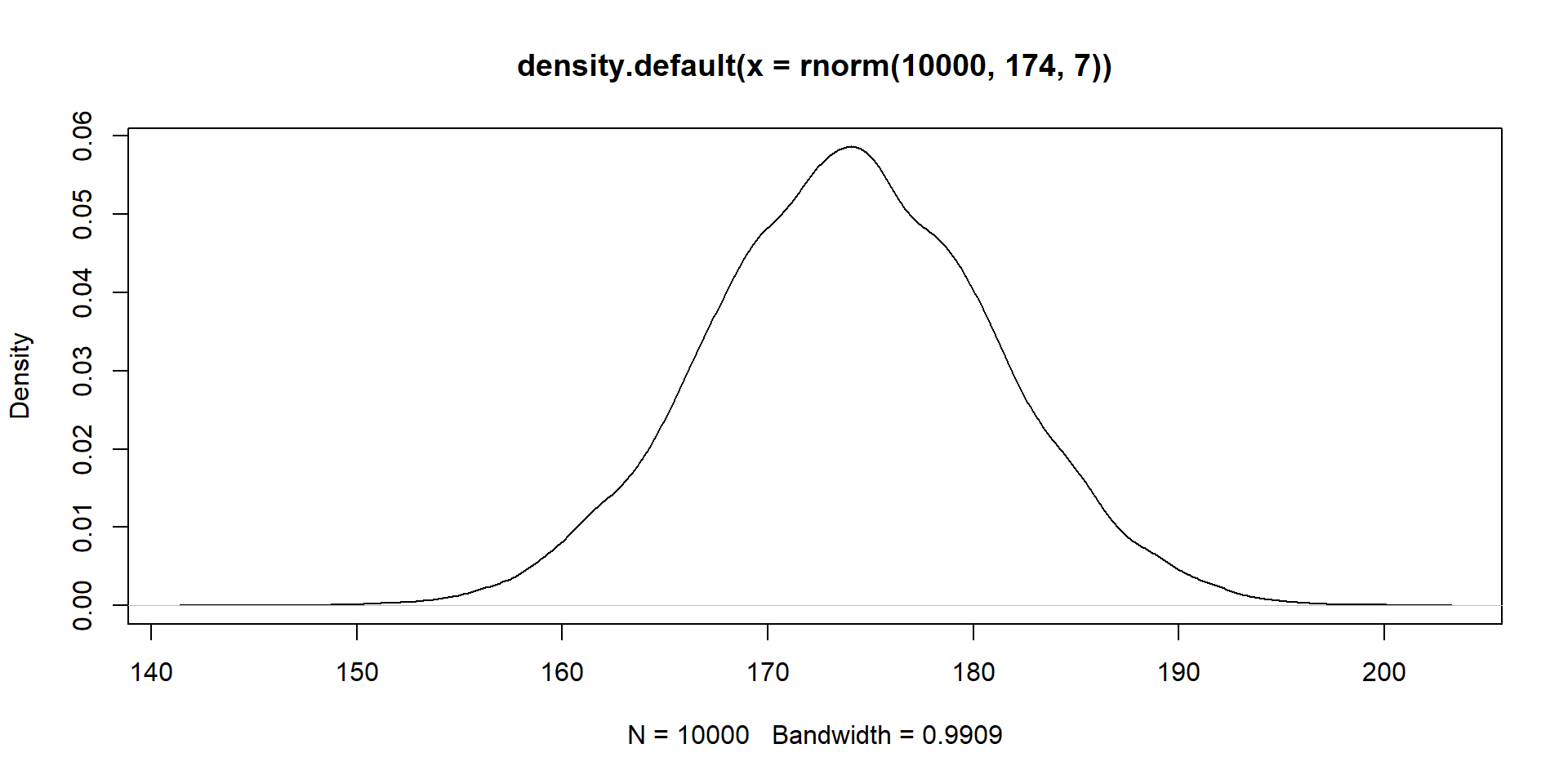
You measure height in a sample (the observable consequences). What’s the data generating process?
We have a (simple) process
\(Normal(\mu, \sigma)\)
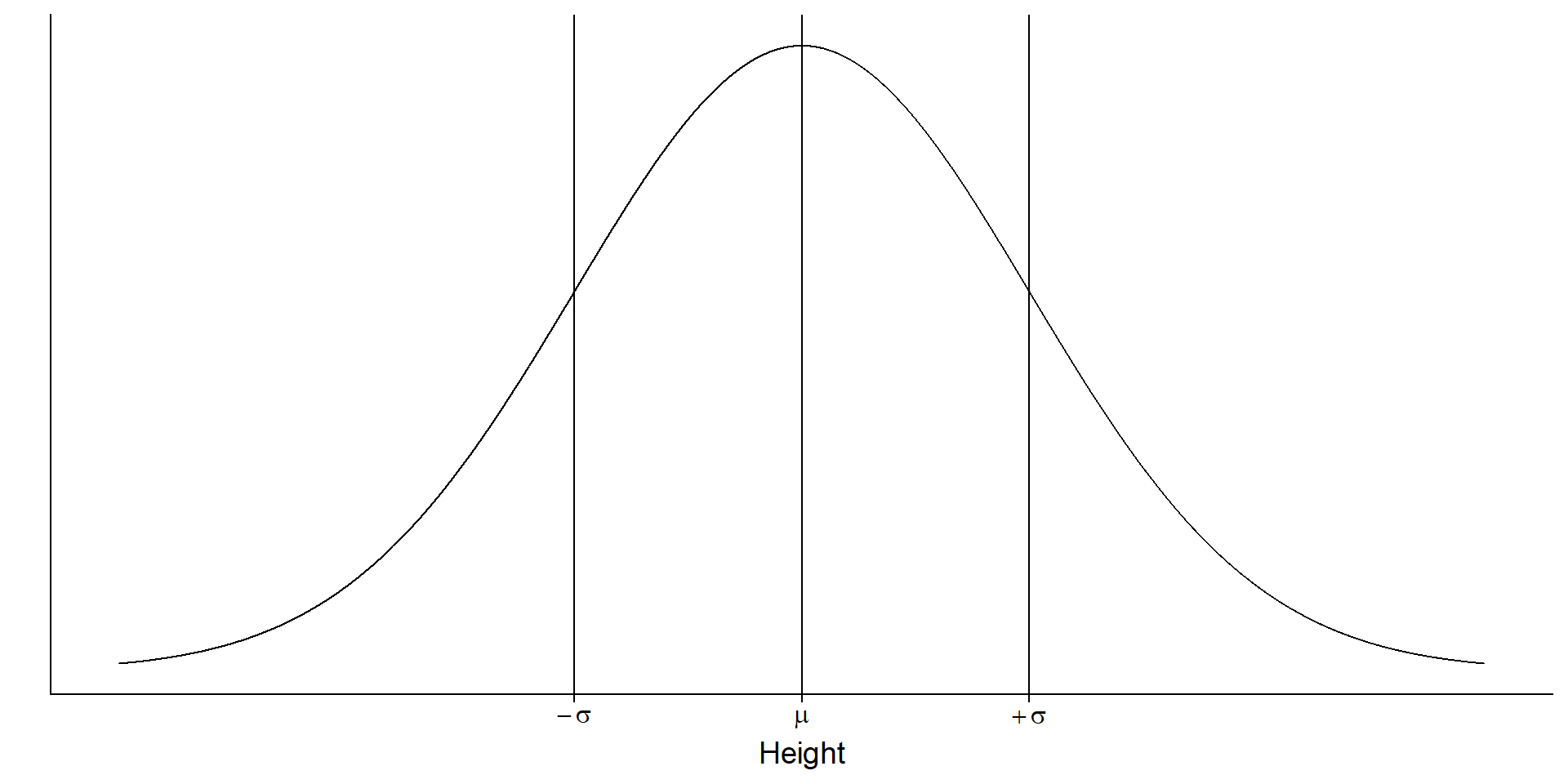
Look familiar?
\(Normal(174, 7)\) aka rnorm(174, 7)
All are processes
rnorm: You say that the underlying data generating process is a normal distributionrbinom: You say that the underlying data generating process is a binomial distribution- You get the idea
Important: Those are assumptions you make explicit and they can be wrong.
Remember our t-tests?
You are explicitly claiming that the observable outcome (the data) have been generated by this true underlying process. The process is a normal distribution with a true effect size (\(\mu\)) and a true standard deviation (\(\sigma\)) for two data generating processes: \(Normal(100, 15)\) and \(Normal(105, 15)\)
Welch Two Sample t-test
data: control and treatment
t = -0.57372, df = 197.92, p-value = 0.5668
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-5.495395 3.018476
sample estimates:
mean of x mean of y
100.3389 101.5774 That’s why simulation is cool
- You’re being explicit about your model of the world
- You translate that model (data generating process) into a concrete formula (code) to create data (the consequences of the data generation process)
- You check how changes to your model influence the data, which inform the conclusions you draw from the data about your model (aka statistical inference)
- No more hiding your model assumptions behind standard software
This all sounds complicated
Here’s the good news: In (much of) the (social) sciences we rely on a common data generating process: the linear model.
t-test, regression, logistic regression, machine learning are all variations of the linear model.
\(y = \beta_0 + \beta_1x\)
All stolen from here
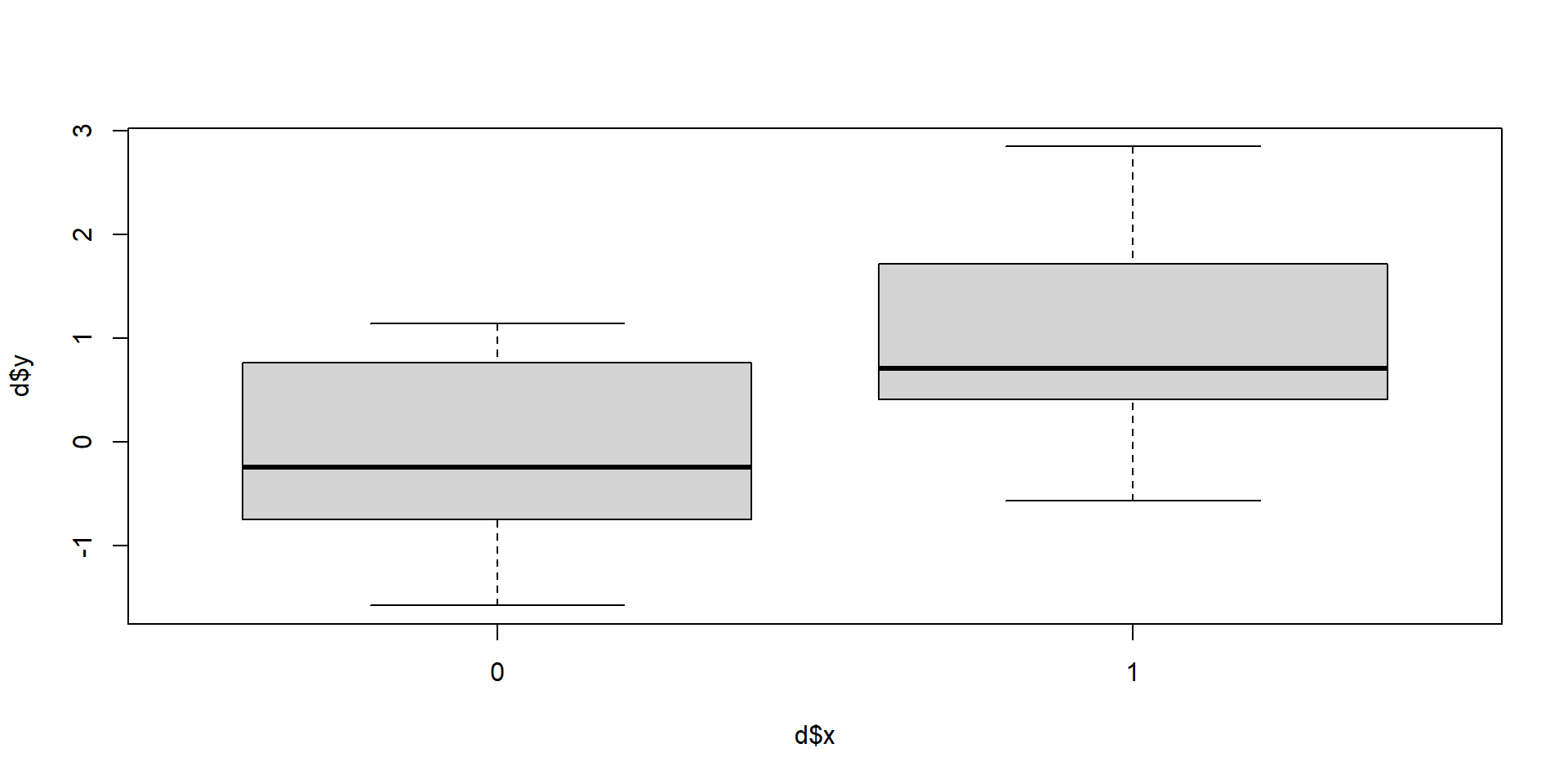
Back to our control and treatment
\(y = \beta_0 + \beta_1x\): What’s \(x\) here?
Group membership
Our group is \(x\).
Finding our inputs again
| group | y | x |
|---|---|---|
| control | -0.2462229 | 0 |
| control | -0.9597997 | 0 |
| control | -0.5238199 | 0 |
| control | 1.1407442 | 0 |
| control | -0.6588351 | 0 |
| control | -1.5772346 | 0 |
| control | 0.6479015 | 0 |
| control | -0.1497161 | 0 |
| control | 1.0852342 | 0 |
| control | 0.4002139 | 0 |
| control | 0.9653700 | 0 |
| control | 0.8700338 | 0 |
| control | -0.8435252 | 0 |
| control | -1.4519022 | 0 |
| control | -0.4569357 | 0 |
| treatment | 2.4476205 | 1 |
| treatment | 0.2285001 | 1 |
| treatment | 0.8593449 | 1 |
| treatment | 1.2349919 | 1 |
| treatment | -0.1423595 | 1 |
| treatment | 0.5735239 | 1 |
| treatment | 0.7055050 | 1 |
| treatment | 2.4935786 | 1 |
| treatment | -0.5713438 | 1 |
| treatment | 0.4377587 | 1 |
| treatment | 2.8445324 | 1 |
| treatment | 1.7562574 | 1 |
| treatment | 0.5085672 | 1 |
| treatment | 0.3798965 | 1 |
| treatment | 1.6759042 | 1 |
What happens when \(x\) is zero?
\[\begin{align} & y = \beta_0 + \beta_1x\\ & y = \beta_0 + \beta_1 \times 0\\ & y = \beta_0 \end{align}\]In other words, when we predict values for the control (aka \(x=0\)) group, our best predictor for outcome scores becomes \(\beta_0\) (also known as the intercept): the mean for the control group.
What happens when \(x\) is one?
\[\begin{align} & y = \beta_0 + \beta_1x\\ & y = \beta_0 + \beta_1 \times 1\\ & y = \beta_0 + \beta_1\\ & y = mean(control) + ? \end{align}\]We already know that the intercept (\(\beta_0\)) is the mean of the control group. How would we now go from that mean to predicting scores of the treatment group? What do we need to add to the mean of the control group to get the mean of the treatment group?
Pictures, please
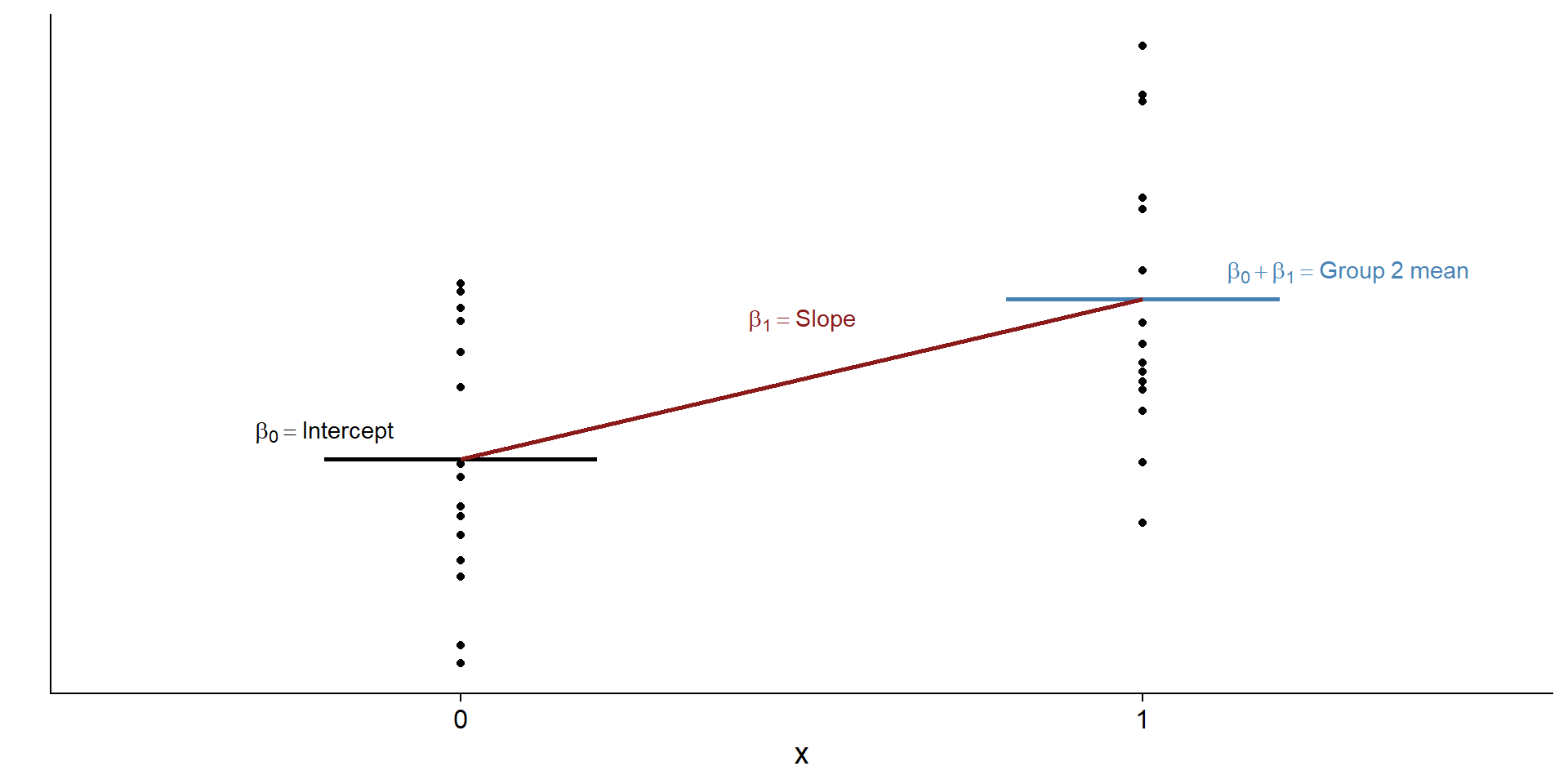
Let’s check that
Mean of the control group (our intercept or \(\beta_0\)):
Difference (our slope or \(\beta_1\)):
Mean of the treatment group (\(\beta_0 + \beta_1\)):
Linear model:
Call:
lm(formula = y ~ x, data = d)
Residuals:
Min 1Q Median 3Q Max
-1.6002 -0.6345 -0.2464 0.7557 1.8157
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.1172 0.2496 -0.470 0.64225
x1 1.1461 0.3530 3.246 0.00303 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.9668 on 28 degrees of freedom
Multiple R-squared: 0.2735, Adjusted R-squared: 0.2475
F-statistic: 10.54 on 1 and 28 DF, p-value: 0.003027So they’re really the same?
Two Sample t-test
data: control and treatment
t = -3.2464, df = 28, p-value = 0.003027
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-1.8691755 -0.4229274
sample estimates:
mean of x mean of y
-0.1172329 1.0288185
Call:
lm(formula = y ~ x, data = d)
Residuals:
Min 1Q Median 3Q Max
-1.6002 -0.6345 -0.2464 0.7557 1.8157
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -0.1172 0.2496 -0.470 0.64225
x1 1.1461 0.3530 3.246 0.00303 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.9668 on 28 degrees of freedom
Multiple R-squared: 0.2735, Adjusted R-squared: 0.2475
F-statistic: 10.54 on 1 and 28 DF, p-value: 0.003027Same logic for a one-sample t-test
There’s no \(x\) here, so all we’re left with is the intercept, which we test against our H0. A single number (aka the mean) predicts \(y\).
\[\begin{align} & y = \beta_0 \end{align}\]Pictures, please

Let’s check that once more
Linear model:
Call:
lm(formula = y ~ 1, data = d[d$x == 1, ])
Residuals:
Min 1Q Median 3Q Max
-1.6002 -0.6200 -0.3233 0.6873 1.8157
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.0288 0.2619 3.929 0.00151 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.014 on 14 degrees of freedomSo they’re really the same?
Call:
lm(formula = y ~ 1, data = d[d$x == 1, ])
Residuals:
Min 1Q Median 3Q Max
-1.6002 -0.6200 -0.3233 0.6873 1.8157
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.0288 0.2619 3.929 0.00151 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.014 on 14 degrees of freedomSame logic for a paired-samples t-test
Remember: Paired samples t-test is just testing the difference in scores against H0. This way, it turns into a one-sample t-test, so all we’re left with is, once again, the intercept, which we test against our H0. A single number (aka the mean) predicts \(y\) (technically \(y_{difference}\)).
\[\begin{align} & y_{treatment} - y_{control} = \beta_0 \end{align}\]Pictures, please

Pictures, please
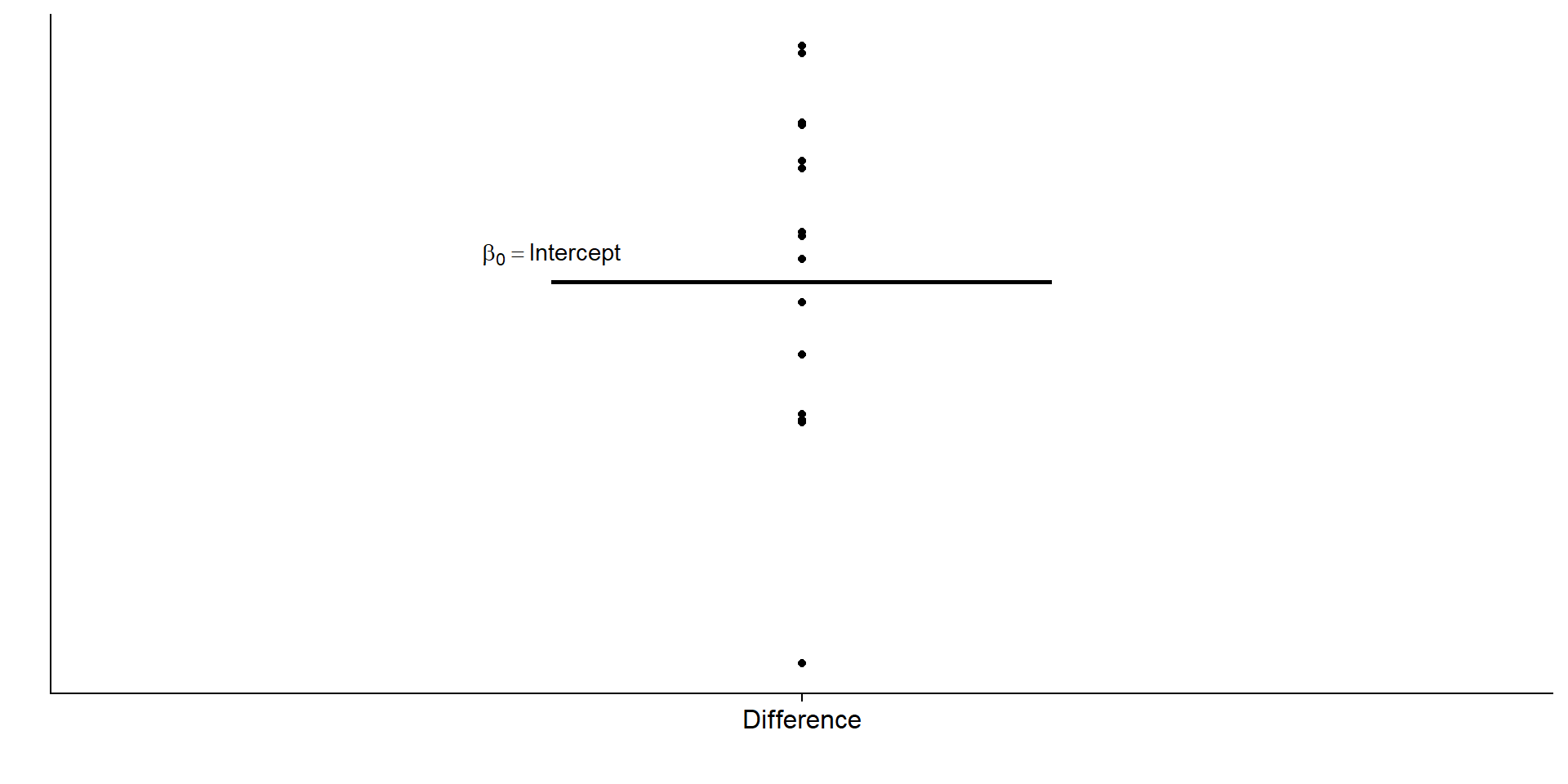
Let’s check that once more
Linear model:
Call:
lm(formula = treatment - control ~ 1)
Residuals:
Min 1Q Median 3Q Max
-2.8026 -0.8407 0.2060 0.8599 1.5478
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.1461 0.3052 3.754 0.00213 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.182 on 14 degrees of freedomSo they’re really the same?
Call:
lm(formula = treatment - control ~ 1)
Residuals:
Min 1Q Median 3Q Max
-2.8026 -0.8407 0.2060 0.8599 1.5478
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 1.1461 0.3052 3.754 0.00213 **
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 1.182 on 14 degrees of freedomWhat about ANOVA then?
Same as before: We predict scores with the group membership (aka the mean in each group). Doesn’t matter whether we predict it from two (t-test) or more groups (ANOVA). Now we just have an indicator for membership for each group: dummy coding.
| groups | \(x_1\) | \(x_2\) |
|---|---|---|
| control | 0 | 0 |
| low | 1 | 0 |
| high | 0 | 1 |
Getting the score for control
| groups | \(x_1\) | \(x_2\) |
|---|---|---|
| control | 0 | 0 |
| low | 1 | 0 |
| high | 0 | 1 |
Getting the score for low
| groups | \(x_1\) | \(x_2\) |
|---|---|---|
| control | 0 | 0 |
| low | 1 | 0 |
| high | 0 | 1 |
Getting the score for high
| groups | \(x_1\) | \(x_2\) |
|---|---|---|
| control | 0 | 0 |
| low | 1 | 0 |
| high | 0 | 1 |
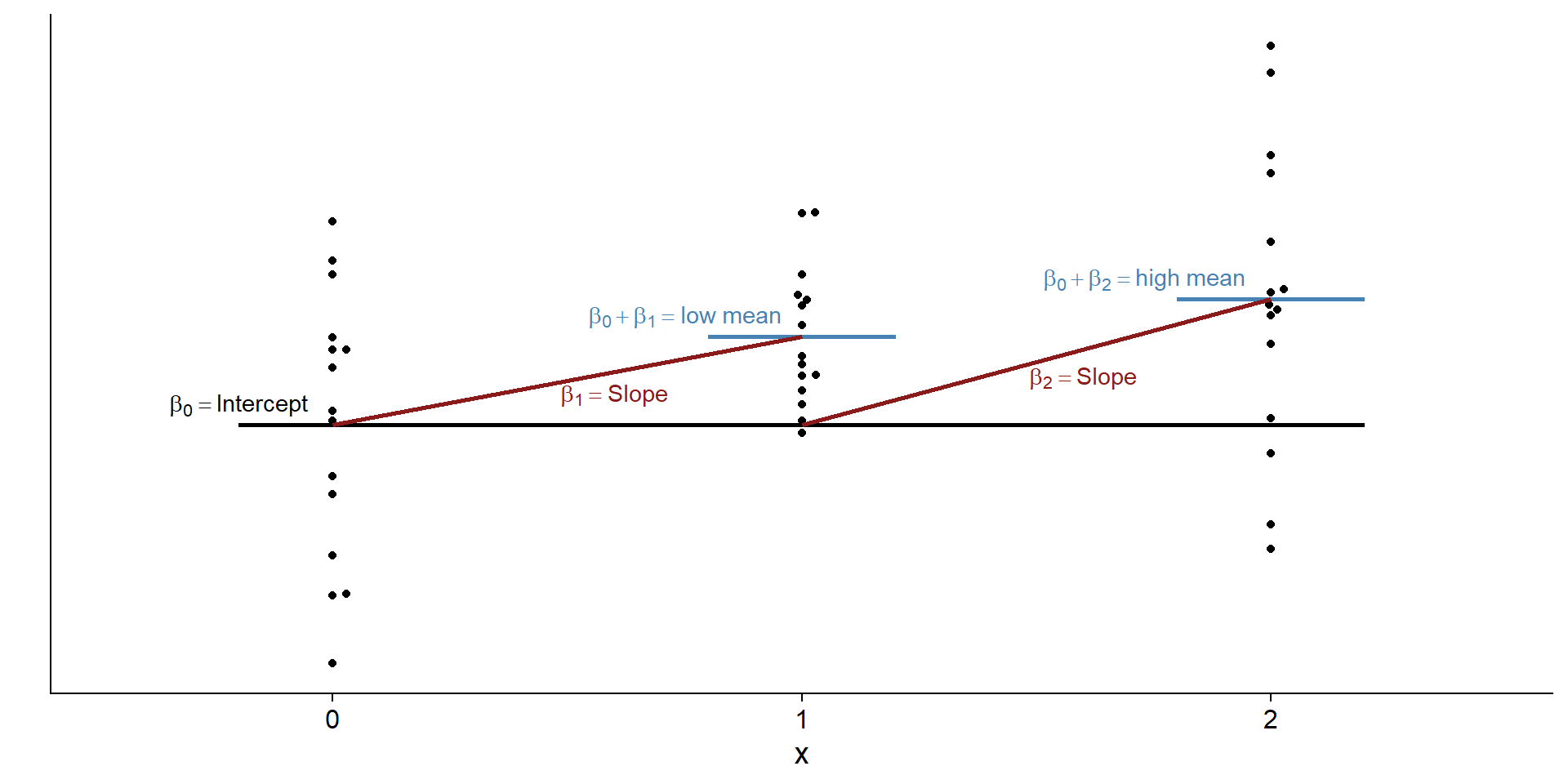
What are \(\beta_1\) and \(\beta_2\) then?
What do we need to go from the control mean to the low mean? What do we need to go from control mean to high mean? Same as with the t-test: the difference between those means.
Pictures, please
Just an extension
If we know how to simulate a t-test, we know how to simulate an ANOVA (because both are just linear models): Imitate the data generating process.
\[\begin{align} & y = \beta_0 + \beta_1x_1 + \beta_2x_2 \end{align}\]Add our dummy codes
y condition group_low group_high
1 106.49227 control 0 0
2 87.82910 control 0 0
3 121.66152 control 0 0
4 93.52831 control 0 0
5 109.83472 control 0 0
6 124.82888 low 1 0
7 108.24242 low 1 0
8 143.63591 low 1 0
9 129.64349 low 1 0
10 121.34641 low 1 0
11 144.14826 high 0 1
12 150.18933 high 0 1
13 141.34749 high 0 1
14 95.10365 high 0 1
15 144.27324 high 0 1Compare to model
# A tibble: 3 × 2
condition mean
<fct> <dbl>
1 control 104.
2 low 126.
3 high 135.
Call:
lm(formula = y ~ group_low + group_high, data = d)
Residuals:
Min 1Q Median 3Q Max
-39.909 -7.267 4.104 9.198 18.096
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 103.869 7.547 13.763 1.04e-08 ***
group_low 21.670 10.673 2.030 0.0651 .
group_high 31.143 10.673 2.918 0.0129 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 16.88 on 12 degrees of freedom
Multiple R-squared: 0.4272, Adjusted R-squared: 0.3317
F-statistic: 4.475 on 2 and 12 DF, p-value: 0.03532Compare to ANOVA
Call:
lm(formula = y ~ group_low + group_high, data = d)
Residuals:
Min 1Q Median 3Q Max
-39.909 -7.267 4.104 9.198 18.096
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 103.869 7.547 13.763 1.04e-08 ***
group_low 21.670 10.673 2.030 0.0651 .
group_high 31.143 10.673 2.918 0.0129 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 16.88 on 12 degrees of freedom
Multiple R-squared: 0.4272, Adjusted R-squared: 0.3317
F-statistic: 4.475 on 2 and 12 DF, p-value: 0.03532No need to use dummies
The lm call will automatically dummy code factors.
Call:
lm(formula = y ~ condition, data = d)
Residuals:
Min 1Q Median 3Q Max
-39.909 -7.267 4.104 9.198 18.096
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 103.869 7.547 13.763 1.04e-08 ***
conditionhigh 31.143 10.673 2.918 0.0129 *
conditionlow 21.670 10.673 2.030 0.0651 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 16.88 on 12 degrees of freedom
Multiple R-squared: 0.4272, Adjusted R-squared: 0.3317
F-statistic: 4.475 on 2 and 12 DF, p-value: 0.03532In a power simulation
n <- 40
m1 <- 100
m2 <- 103
m3 <- 105
sd <- 8
draws <- 1e4
pvalues <- NULL
for (i in 1:n) {
group1 <- rnorm(n, m1, sd)
group2 <- rnorm(n, m2, sd)
group3 <- rnorm(n, m3, sd)
dat <- data.frame(
scores = c(group1, group2, group3),
condition = as.factor(rep(c("group1", "group2", "group3"), each = n))
)
m <- summary(lm(scores ~ condition, data = dat))
pvalues[i] <- broom::glance(m)$p.value
}
sum(pvalues < 0.05) / length(pvalues)[1] 0.7Getting that p-value
You can access the p-value by storing the summary in an object (as a list) and accessing its component. For the lm summary, that’s a bit less straightforward. See https://stackoverflow.com/questions/5587676/pull-out-p-values-and-r-squared-from-a-linear-regression.
About that effect size
Let’s just swap the low and high conditions.
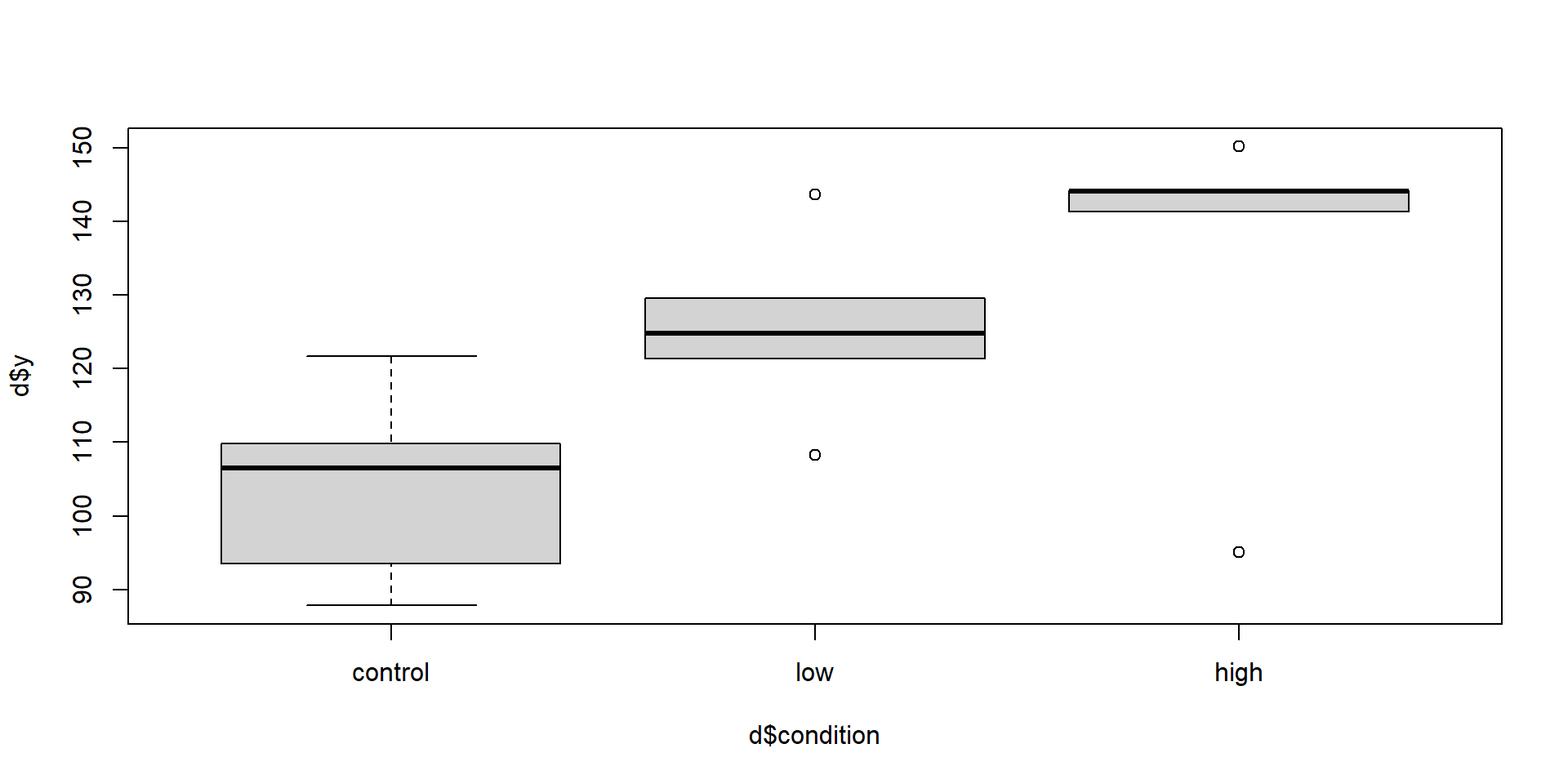
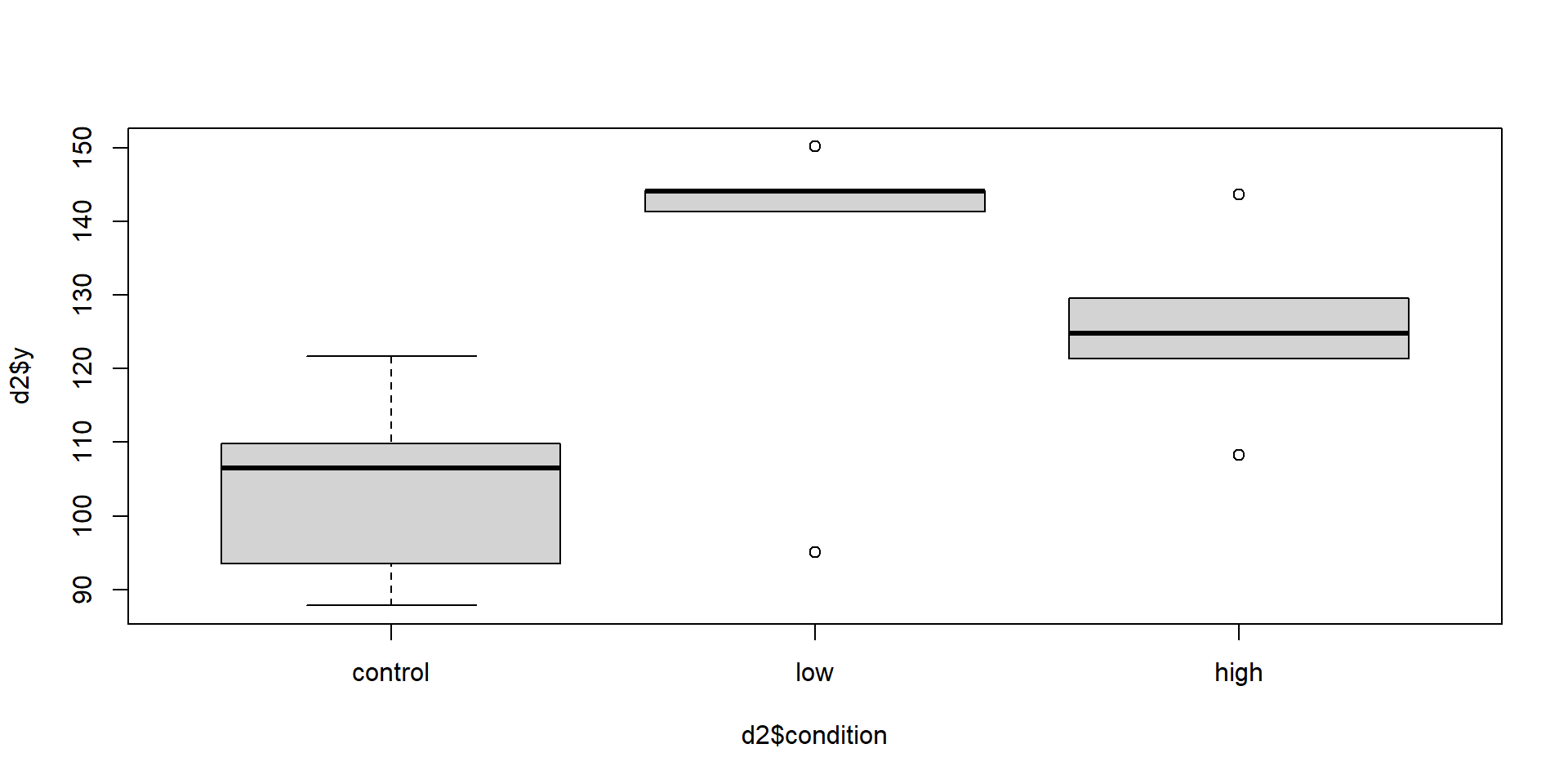
Now what are the model statistics?
Call:
lm(formula = y ~ condition, data = d)
Residuals:
Min 1Q Median 3Q Max
-39.909 -7.267 4.104 9.198 18.096
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 103.869 7.547 13.763 1.04e-08 ***
conditionlow 21.670 10.673 2.030 0.0651 .
conditionhigh 31.143 10.673 2.918 0.0129 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 16.88 on 12 degrees of freedom
Multiple R-squared: 0.4272, Adjusted R-squared: 0.3317
F-statistic: 4.475 on 2 and 12 DF, p-value: 0.03532
Call:
lm(formula = y ~ condition, data = d2)
Residuals:
Min 1Q Median 3Q Max
-39.909 -7.267 4.104 9.198 18.096
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 103.869 7.547 13.763 1.04e-08 ***
conditionlow 31.143 10.673 2.918 0.0129 *
conditionhigh 21.670 10.673 2.030 0.0651 .
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 16.88 on 12 degrees of freedom
Multiple R-squared: 0.4272, Adjusted R-squared: 0.3317
F-statistic: 4.475 on 2 and 12 DF, p-value: 0.03532What about the effect size?
# Effect Size for ANOVA
Parameter | Eta2 | 95% CI
-------------------------------
condition | 0.43 | [0.03, 1.00]
- One-sided CIs: upper bound fixed at [1.00].# Effect Size for ANOVA
Parameter | Eta2 | 95% CI
-------------------------------
condition | 0.43 | [0.03, 1.00]
- One-sided CIs: upper bound fixed at [1.00].How would I go for power now?
- Back to standardized vs. unstandardized
- Best to “draw” first
- Determine contrast of interest
What about correlated measures?
Remember drawing two scores from the same unit?
\[ \begin{bmatrix} var & cov \\ cov & var \\ \end{bmatrix} \]
We just need to extend that to the number of measures:
\[ \begin{bmatrix} var & cov & cov\\ cov & var & cov\\ cov & cov & var \end{bmatrix} \]
What we need to know
- The standard deviation for each measure (\(\sigma_{x_1}\))
- The correlation between each measure (\(r_{x_1x_2}\))
\[ \begin{bmatrix} SD_{x_1} & r_{x_1x_2} & r_{x_1x_3}\\ r_{x_2x_1} & SD_{x_2} & r_{x_2x_3}\\ r_{x_3x_1} & r_{x_3x_2} & SD_{x_3} \end{bmatrix} \]
Same as before
Define our parameters and get variance-covariance matrix.
means <- c(control = 100, low = 103, high = 105)
sd <- 8
correlation <- 0.4
covariance <- correlation * sd * sd
our_matrix <- matrix(
c(
sd**2, covariance, covariance,
covariance, sd**2, covariance,
covariance, covariance, sd**2
),
ncol = 3
)
our_matrix [,1] [,2] [,3]
[1,] 64.0 25.6 25.6
[2,] 25.6 64.0 25.6
[3,] 25.6 25.6 64.0Then draw from multivariate normal
library(MASS)
set.seed(42)
d <-
mvrnorm(
200,
means,
our_matrix
)
d <- as.data.frame(d)
d$id <- factor(1:200)
head(d) control low high id
1 102.65124 93.16169 86.70051 1
2 98.88458 109.62849 109.98486 2
3 94.79098 97.77544 108.68290 3
4 90.66920 94.06642 111.49926 4
5 98.78622 106.29663 95.40168 5
6 99.54111 110.82243 99.60934 6Let’s check
Can we recover our numbers of means = c(100, 103, 105), SD of 8, and correlation of 0.4?
We can’t use lm this time
The linear model has several assumptions, one of which is that observations are independent. They clearly aren’t (we specified a correlation between them after all). So we need to go for models that take this dependence into account.
For most cases, data need to be in the long format:
Long format
| control | low | high | id |
|---|---|---|---|
| 102.65124 | 93.16169 | 86.70051 | 1 |
| 98.88458 | 109.62849 | 109.98486 | 2 |
| 94.79098 | 97.77544 | 108.68290 | 3 |
| 90.66920 | 94.06642 | 111.49926 | 4 |
| 98.78622 | 106.29663 | 95.40168 | 5 |
| 99.54111 | 110.82243 | 99.60934 | 6 |
| id | condition | score |
|---|---|---|
| 1 | control | 102.65124 |
| 1 | low | 93.16169 |
| 1 | high | 86.70051 |
| 2 | control | 98.88458 |
| 2 | low | 109.62849 |
| 2 | high | 109.98486 |
Let’s run the RM ANOVA
Error: id
Df Sum Sq Mean Sq F value Pr(>F)
Residuals 199 21774 109.4
Error: Within
Df Sum Sq Mean Sq F value Pr(>F)
condition 2 2927 1464 38.48 5.25e-16 ***
Residuals 398 15135 38
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1Just a (hierarchical) linear model
Check here for more background info.
This is the max
The workshop won’t go further than (interactions) with repeated measures. More resources on simulating more complicated designs at the final wrap up.
Takeaways
- Understand the logic behind the data generating process
- See how the linear model is our data generating process
- Apply this to a setting with multiple categories in a predictor